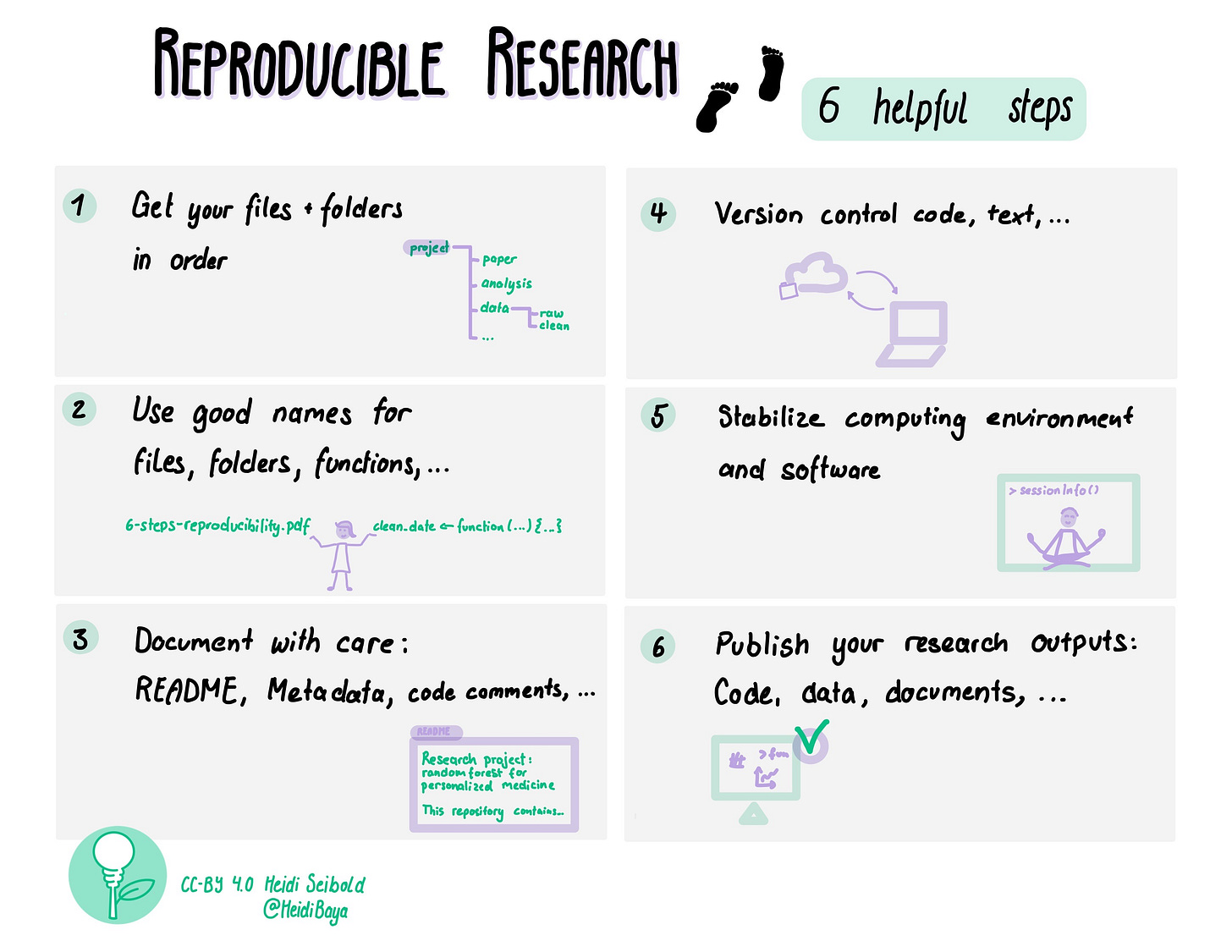
Document with care: README, Metadata, code comments, … to make both others and future you happy 😃
E-mail course: 6 steps towards reproducible research (step 3)
Hello friend 👋,
welcome back! In this newsletter we will discuss step 3 of our 6 steps towards reproducible research: Document with care: README, Metadata, code comments, …
What parts of my research project need documenting?
That’s for you to decide. There is no super-clear catch all answer. Here are a few thoughts from my side though 😉
README
One thing that I always do is to add a README-Text-File to each project. In the README I write the most important info about the project: What is it about? Who is involved? Where to find files? How to cite it? Where to find the paper? …
Code documentation
In my research projects code plays an important role. That might be different for you → feel free to skip.
To make my code as understandable as possible for others, I use literate programming (mixing text and code to make it easier to read, e.g. RMarkdown) or add clear code comments. When writing functions in R I additionally use the standardised way to document R functions (via Roxygen2).
An example of code comments in R (“#”):
## Load package + data
library("model4you")
data("MathExam14W", package = "psychotools")
## scale points achieved to [0, 100] percent
MathExam14W$tests <- 100 * MathExam14W$tests/26
MathExam14W$pcorrect <- 100 * MathExam14W$nsolved/13
## select variables to be used
MathExam <- MathExam14W[ , c("pcorrect", "group", "tests", "study",
"attempt", "semester", "gender")]Metadata
Metadata is information about your data. It’s information on the license of the data, who owns it, what information the data cointain, …
Many research fields have standards for metadata. If you can’t find one for your field you can use a common standard (e.g. Dublin Core) or just ask a data manager or librarian at your institution. You can write metadata similar to a README (see e.g. this guide from Cornell University). If you upload your data to a data platform (e.g. Dryad) you won’t have to think about it as the platform usually takes care of that (Dryad uses Dublin Core).
Other
Whatever you work on, there might be parts of your research project that are difficult to understand. Say you work in a lab, then your documentation is a lab notebook. Or you do interviews, then your documentation may be your interview strategy. Anything that might be useful for others is worth keeping and worth sharing. After all, we all want to build on the work of others in order to make the world a little better.
Your tasks ✅
Check if your current research project already has a README. If not, create one 🙌
Do you write code? Make a habit of writing code comments right when you create the code.
Will be coding this upcoming week? Start doing it (if you don’t already 😉).
Won’t be coding this week? Go to a recent script and check if you did a good job. If not, try code comments 💪.
Check out the literature linked in this newsletter issue. Anything in particular you find interesting? Share your newly gained knowledge with your peers 🤓🤓🤓.
Further reading
Want to learn more? Check out:
Landing Page - README file, The Turing Way
A beginner’s guide to writing documentation, Write The Docs
R Markdown: The Definitive Guide, Yihui Xie, J. J. Allaire, Garrett Grolemund
knitr - Elegant, flexible, and fast dynamic report generation with R, Yihui Xie
Guide to writing "readme" style metadata, research data management service group, Cornell University
Any questions remaining? Feel free to ask me!
Feel like you’re in need of some time off the daily grind to wrap your head around all this stuff? Or you want to meet others who are into Open Science too?
Come join us at the Open Science Retreat!

Next time we’ll discuss step 4: Version Control. Oh - that is a topic I love, so stay tuned 💜
Cheers,
Heidi